Researchers from Washington have developed a machine learning algorithm that assists in generating a 3D model of cells from a partial set of 2D images
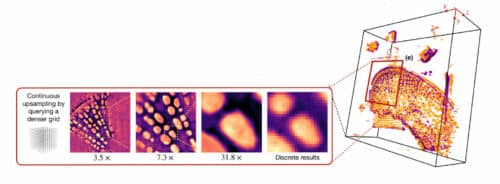
It was difficult to understand and study the details of cells from 2D images that consisted of very restricted information. Hence, researchers from the Mckelvey School of Engineering have invented a machine learning algorithm to obtain a detailed 3D model of cells from a set of 2D images that consists of partial information, obtained using conventional microscopy tools available in many labs even today. The essential element for this research was a neural field network, a kind of machine learning system that learns a mapping from spatial coordinates to the corresponding physical quantities. After training completion, researchers may test by pointing to any coordinate for which the model can provide the image value.
“We train the model on the set of digital images to obtain a continuous representation,” said Ulugbek Kamilov, assistant professor of electrical and systems engineering and computer science and engineering. “Now, I can show it anyway I want. I can zoom in smoothly and there is no pixelation.”
The main attribute of neural field networks is they are not required to be trained on a large amount of similar data. It only needs a sufficient amount of 2D images of the sample; hence, it can represent its entirety, inside and out. The network is trained using an image similar to any other microscopy image. The process goes like this, the cell is enlightened from below, and the light travels through it and is captured from another side, this creates an image. Now, the network extracts its best shot at recreating that structure. If the output is wrong, the network is tweaked. If it’s correct, that pathway is reinforced. Once the predictions match real-world measurements, the network is ready to fill in parts of the cell that were not captured by the original 2D images.
The imaging system can zoom in on a pixelated image and fill in the missing pieces, creating a continuous 3D representation. “Because I have some views of the cell, I can use those images to train the model,” Kamilov said. This is done by feeding the model information about a point in the sample where the image captured some of the internal structure of the cell.
The above research provides a model and an easy-to-store and true representation of the cell. This can be more useful than the real thing.
Click here for the Published Research Paper







