Currently, our entire planet is grappling with a calamity of a magnitude never ever experienced before—the deadly and highly infectious Covid-19 pandemic. It is a ‘black swan event’ comparable with the World War II catastrophe that adversely affected every aspect of human life
The world has suffered under the influence of various epidemics in the past. The Severe Acute Respiratory Syndrome (SARS) virus, which was traced to China in 2002, infected over 8,000 people and killed approximately 800 people across 29 countries. In March 2014, the Ebola epidemic, which originated in West Africa, killed more than 11,000 people in six countries. Pandemics, such as Covid-19, currently affecting the entire world, have never been seen earlier. This highly infectious disease, spreads mainly via contact with an infected person or by touching a surface having the virus on it.
As per the World Health Organization (WHO), the Covid-19 pandemic had infected a staggering 27.49 million people, causing as many as 0.89 million deaths in 216 countries by September 9, 2020. Governments had to seal borders, impose travel limitations, and shut down educational institutions, market places, and public places, giving rise to an economic recession. According to the United Nations, more than 1,200 million wage earners suffered due to the shutting down of workplaces and stalled manufacturing. No effective treatment is available as yet to cure or prevent the disease. However, doctors and scientists are struggling to find an effective solution.
Several research projects and clinical trials are going on in many countries. These efforts require extensive research spanning areas like bioinformatics, epidemiology, and molecular modelling. A massive IT support and computational capacity are needed to execute these complex research programmes. With the Covid-19 cases rising rapidly, a wealth of data is available for modelling and analysis. Supercomputers, with their very high processing power, are most suited for performing such functions. The spikes present on the surface of the Covid-19 invade cells in the human body. Supercomputers help look through the databases of existing antiviral drug compounds to help find a drug that could bind with these spikes, inhibiting the virus from infecting humans. Cloud computing platforms assist researchers to access the data and the apps needed.

Given that 216 countries are affected, rapid data sharing is critical for understanding the origins, spread of the infection, its prevention, treatment, and care. The IT platforms make low-cost dissemination and collaboration of data possible. Deep learning models of artificial intelligence (AI) make accurate detection of Covid-19 possible and help differentiate it from community-acquired pneumonia and lung diseases. Bioinformatics and geographic information systems (GIS) integrate computer science, IT, and biology towards developing new algorithms and creating tools for the analysis and management of diverse types of information to evaluate relationships between large data sets.
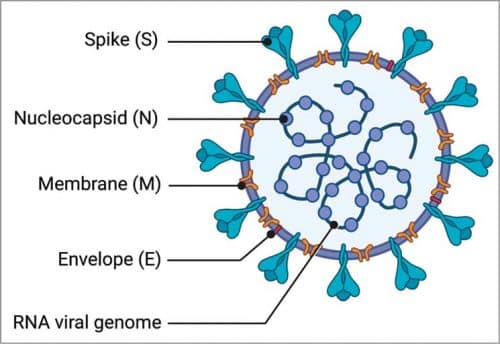
Structure of Covid-19
The source of this unknown respiratory infection is related to a novel coronavirus that caused outbreaks of SARS from 2002 to 2004. The virus is called SARS-CoV-2, and the World Health Organization has named it Covid-19 (coronavirus detected in Dec 2019). Covid-19 particles are spherical in shape and have protein spikes protruding from the surface. These spikes latch onto human cells and undergo a structural change such that the viral membrane gets fused with the human cell membrane. The viral genes then enter the host cell producing more viruses. Covid-19 has four structural proteins, envelope (E), spike (S), membrane (M), and nucleocapsid (N).
The S, M, and E proteins form the envelope of the virus. The S-proteins give the virus a crown-like appearance, thus the name coronavirus. The M protein, the most abundant, is responsible for the shape of the envelope. The E protein, the smallest, is the structural protein. Both the S and M proteins help in virus assembly during replication. N protein is largely involved in the replication cycle (assembly and budding) and the host cellular response to viral infection. The Covid-19 spikes, which are ten to twenty times more likely to bind human cells compared to the 2002 SARS virus spikes, enable it to spread more easily from person to person.

In spite of similarities in the sequence and structure of the spikes of the two viruses, the 2002 SARS virus antibodies do not successfully bind to the Covid-19 spike protein, indicating that the vaccine and antibody-based treatments have to be unique to this new virus. Researchers are now experimenting with the spike protein to isolate antibodies from people who have recovered from infection to treat new infections before a vaccine becomes available. Cultured cells are used by scientists to make large quantities of the protein available for analysis. Cryo-electron microscopy is used to take pictures of the spike protein, and the images are combined to construct a 3D view of the virus.
Application of IT in fighting Covid-19
Supercomputers
The scientific community needs to study how the proteins’ spikes attach to the wall of a human cell, how does the RNA of the virus get inside the human cell, and how does it take over the human DNA machinery. Large numbers of these experiments, when conducted in the lab, take a long time to be completed.
Simulations of experiments using supercomputers help biologists and scientists to find treatments and vaccines in the shortest time possible. The present-day supercomputers function at a speed as high as ‘peta’—a level higher than ‘tera’ (the levels in ascending order are kilo, mega, giga, tera, peta, and exa). These are a powerful resource for simulation of biochemical processes, materials, and chemical agents and help process a massive number of calculations as well as solve a vast number of complex and complicated equations related to bioinformatics, epidemiology, and molecular modelling. As the Covid-19 cases are increasing at a very high rate, a huge amount of data is available to researchers to find a drug that could work against the novel coronavirus.
Supercomputers look through the databases of existing drug compounds to identify drug compounds and molecules that have the highest binding affinity with the virus to disarm it most efficiently. These supercomputers also study the structure of the virus and how it interacts with cells in the human body as well as analyse the spread of the virus in a population.
In a global effort to tackle the pandemic, a number of countries are employing supercomputers to expedite their research programmes. A joint public-private effort, which includes IBM, Google, Amazon, MIT, Carnegie Mellon, and NASA called ‘Covid-19 High-Performance Computing Consortium,’ has been launched in the US. Japan is using the Fugaku supercomputer (successor to the ‘K’ computer, which had the highest calculation speed in the world).
China is reported to be using the Tianhe-1 supercomputer to diagnose Covid-19 patients from chest scans. In India, the Centre for Development of Advanced Computing (C-DAC) is collaborating with the National Institute of Virology (NIV), IITs, Indian Institutes of Science Education and Research (IISER), labs under the Council of Scientific and Industrial Research (CSIR), Department of Biotechnology (DBT), as well as industries and startups for drug repurposing simulations towards discovering a suitable and effective drug for Covid-19. C-DAC, a part of the Indian Institute of Science (IISc), has launched a hackathon SAMHAR-Covid-19 to fight the coronavirus. (SAMHAR stands for ‘Supercomputing using AI, ML, Healthcare Analytics based Research, and it means ‘to vanquish’ in Sanskrit.).
The hackathon is open to researchers, academicians, micro small and medium enterprises (MSMEs), startups, and industries to bring out innovative and implementable ideas for predicting, forecasting, and building healthcare models as well as facilitating drug discovery using supercomputers and AI technology.
In India, work on supercomputers started in 1987 under the erstwhile C-DoT (Centre for Development of Telecommunication). Today, the country has two supercomputers and ranks 22nd in the supercomputing power (China tops the list with 228 systems). It is interesting to note that the first supercomputer assembled indigenously, Param Shivay, was installed in IIT (BHU) in February 2019. Similar systems, Param Shakti and Param Brahma, were installed at IIT-Kharagpur and IISER, Pune, respectively.
Cloud computing
Usually, the IT infrastructure in hospitals and labs is limited to an on-premise server with a limited data storage capacity, restricting the amount of memory and processing resources and the number of applications that can be used. To scale up the capacity, new equipment has to be added. Cloud technology increases the processing capacity dynamically and quickly without the need to procure new equipment and makes utilisation of a variety of cloud applications possible that are otherwise cost-prohibitive to purchase.
Cloud computing provides the capability to shift from on-premises data solutions to colocation or cloud-based services. The distributed nature of cloud technology makes it accessible to multiple organisations as the data and applications stored in a cloud network can be accessed by anyone who has the right to access it. This permits transparency and data sharing within a secure framework.
The ability to share and move data easily provides interoperability and rapid knowledge sharing. Cloud computing enables healthcare professionals working in the field to quickly and easily transmit on-site data to a shared location, which can be accessed instantly by the researchers. Leading cloud computing organisations have been proactively supporting cloud-based healthcare research. The US Centres for Disease Control and Prevention (CDC) used cloud technology to combat influenza in the wake of the 2009 H1N1 outbreak and also found it highly effective for viral research during West Africa’s Ebola crisis. IBM has now made available a number of cloud computing resources for free to Covid-19 researchers to model the spread of the disease and identify drugs for effective treatments. Amazon Web Services (AWS) has provided twenty million dollars in cloud credits to subsidise research into diagnostic tools related to Covid-19.
Oracle is actively participating in coronavirus vaccine development by providing computing platforms for clinical trials. Oracle has also introduced Oracle Clinical Trials Systems to gather data on Covid-19 drug testing and therapeutic learning to serve as a repository for all Covid-19 treatments being administered.
Artificial intelligence and heuristic methods
Artificial intelligence (AI) and heuristic methods, in particular, machine learning, data mining, cluster analysis, pattern recognition, and knowledge representation, are helping meet the challenges posed by the Covid-19 pandemic. AI helps in reducing the workload of healthcare workers by tracking, screening, early detection, and diagnosis of the infection. It helps researchers track the spread of the virus and predicts mortality risk by analysing the data of the patients.
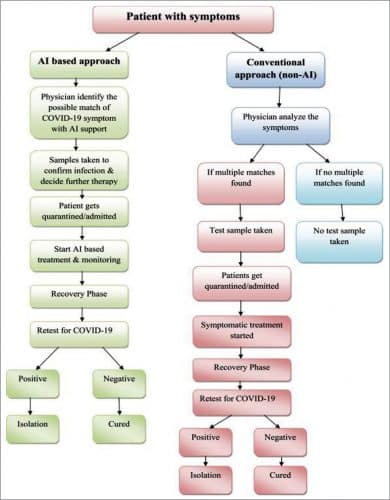
AI helps in identifying high-risk patients by population screening, providing medical help, making notifications, and controlling the infection. The flow diagram on next page compares the AI and non-AI based procedures followed to identify the Covid-19. The AI-based procedure provides higher accuracy and reduces complexity, time taken, and the number of steps needed.
Key areas where the AI approach has been found encouraging and fruitful are knowledge acquisition from genetic data, interpretation of nucleotide sequences, prediction of protein’s structure and function, and drug design. AI promptly analyses the irregular symptoms and other ‘red flags’ present in patients to help healthcare functionaries make faster and cost-effective decisions. It helps in identifying the clusters and ‘hot spots’ of infection, predicting the future course of the disease and chances for its reappearance, contact tracing of the infected individuals, and providing appropriate treatments.
Application of GIS to Covid-19
Geographic understanding is essential in detecting, understanding, and responding to pandemics such as Covid-19. A geographic information system (GIS) is a computer system for capturing, storing, checking, and displaying data related to positions on the earth’s surface in a meaningful way through interactive maps or other infographics. GIS techniques and tools are useful in finding and monitoring large-scale population-based occurrences such as disease clusters, outbreaks of infection, and possible associations between infections and environmental factors.
GIS helps public health agencies, policymakers, and administrators to map disease occurrence against multiple parameters such as demographics, environment, geographies, and past occurrences to understand the origin of an outbreak, its spread pattern, and intensity. GIS enables identifying at-risk populations, planning targeted intervention, evaluating facilities and healthcare capacities, and establishing effective communication among supporting agencies to ensure a coordinated response.
It helps epidemiologists study the outbreak of diseases and how the incidence of congenital diseases varies over space and time. WHO, Red Cross, academics, local governments, and hospitals employ GIS to communicate spatial data sets to the public and help policymakers to prepare and allocate resources appropriately.
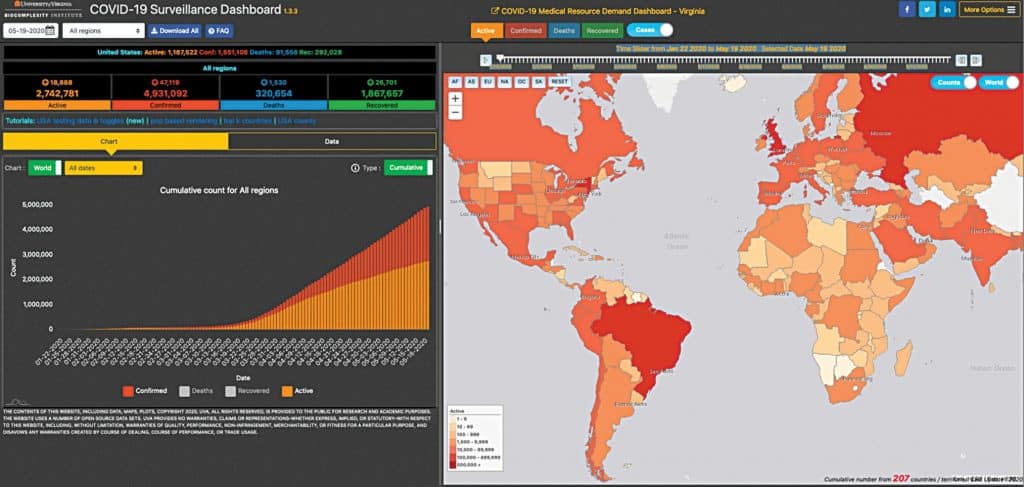
The GIS dashboard app, created by John Hopkins University, Baltimore, allows users to click on a country to identify confirmed cases and deaths. Another app, Covid-19 Surveillance Dashboard, created by the University of Virginia, provides a visualisation of Covid-19 cases, recoveries, and deaths across the globe on a real-time basis. Prediction and forecasting systems allow users to identify areas where shortages in medical equipment are likely to reach crisis point and predict surges in the cases to enable better planning for taking pre-emptive actions so that hospitals do not run out of beds, ventilators, masks, and other supplies.
Some more technologies being used to fight Covid-19
Blockchain
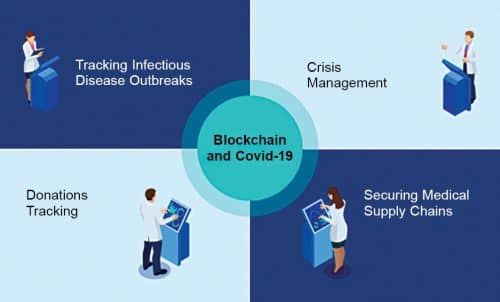
As Covid-19 is highly infectious, there is an urgent need for speeding up the detection of virus carriers and halting their spread. Hence, blockchain technology has emerged as a key technology in the critical domain of pandemic management. Blockchain technology provides a structure to store the transactional records in several databases in a network connected through peer-to-peer nodes, referred to as a ‘digital ledger.’ Every transaction in this ledger is authenticated by the unique digital signature of the owner, safeguarding it from tampering and making the information highly secure.
Blockchain makes it possible for anyone to see the data but not tamper with it. Blockchain applications provide robust, transparent, secure, and cheap means for storing sensitive patient data and facilitating effective decision-making. Healthcare organisations can create a centralised database and share the information with only the appropriately authorised people. This technology has the potential of becoming an integral part of the global response to the coronavirus pandemic by tracking the spread of the disease, maintaining the sustainability of medical supply chains, and managing insurance payments. Its decentralised nature and cryptographic algorithm make it immune to attack and hacking.
Open source technologies
During epidemic outbreaks, rapid data sharing is critical to allow for a greater understanding of the origins and spread of the virus and function as a source for effective prevention, treatment, and care. The capacity of information technologies permits low-cost dissemination and collaboration of data, which enables the creation of a large number of repositories and information technology platforms for data sharing. A substantial number of these activities are coordinated by international organisations such as the World Health Organization (WHO) and European Centre for Disease Prevention and Control. An increasing number of bottom-up, open-data enterprises and open-source projects have been initiated, enabling easy and quick access to research data and scientific publications as well as sharing designs for the production of critical medical equipment such as ventilators and face shields.
3D printing
The existing healthcare system capacity in various countries, planned for the normal times, is limited and unable to meet the rapidly increasing demand for medical hardware such as face masks, ventilators, and breathing filters, etc, required to treat Covid-19 patients. Governments around the world are taking increasingly radical actions to increase production and enhance the supply of the necessary medical equipment. Three-dimensional (3D) printing technology can be of immense use during this emergency in supporting the endeavour of healthcare workers and hospitals in keeping patients alive.

Nanotechnology
The Covid-19 outbreak has tested the limits of healthcare systems, posing serious questions about the adequacy of conventional therapies and diagnostic tools. Quarantine, isolation, and infection-control measures are being used to prevent the spread of the virus and isolate those who show the symptoms.
However, a specific antiviral agent is lacking to treat the infected and decrease viral spreading and transmission. The use of nanotechnology offers new opportunities for the prevention, diagnosis, and treatment of Covid-19. Nanotechnology is broadly defined as the design and application of materials and devices where at least one dimension is less than a hundred nanometres.

The application of nanotechnology in the medical field called ‘nanomedicine’ includes the use of nanomaterials for diagnosis, treatment, control, and prevention of diseases.
Nanoparticles can be extensively used in the development of better and safer drugs, tissue-targeted treatments, personalised nanomedicines, and early diagnosis as these have unique properties such as small size, improved solubility, surface adaptability, and multi-functionality.
Nanotechnology can help fight against Covid-19 by offering infection-safe personal protective equipment (PPE) to enhance the safety of healthcare workers and develop effective antiviral disinfectants and surface coatings to inactivate the virus and prevent its spread.
Nano-based biosensors could be used in diagnostics for viral infection with high specificity and sensitivity. Nanoparticle-based markers can be used to study the mechanism by which viruses infect host cells. A new generation of vaccines could be developed based on nanomaterials with improved antigen stability, target delivery, and controlled release.
Contaminated surfaces in public places, such as hospitals, parks, public transportation, and schools, are a common source for outbreaks of infection. Nano-based surface coatings have the potential for the prevention of such infections.
Drones and robots
Drones are being deployed for disinfection and street patrols for food and medicine delivery in the quarantined areas to contain the spread of the novel coronavirus. China was the first to adapt and co-opt drones to enforce the world’s largest quarantine exercise. The drone’s software can be modified to enforce preventive measures and increase infection detection and crowd management, reducing the risks of widespread surveillance and law enforcement.
Robots are another new technology being used to control the spread of Covid-19. These have been used to provide services and care for those quarantined or practising social distancing right from the time of the initial outbreak of the pandemic in China to its spread across the globe. Scientists and engineers have fast-tracked the ‘testing’ of robots and drones in public. Robots will help all the agencies who are seeking the most expedient and safest way to grapple with the outbreak and limit its further spread.

Future aspects
Healthcare professionals and medical scientists around the world are racing to gather and analyse the massive amounts of data in an effort to develop vaccines and treatments to bring the pandemic to an end. To fight the Covid-19 pandemic, healthcare delivery systems need the support of technologies such as supercomputers, cloud computing, artificial intelligence, machine learning, and geographic information system. Biotech as well as pharmaceutical organisations need extensive software support in terms of writing algorithm programs, developing software systems, and managing databases to develop suitable treatments and vaccines.
Healthcare organisations need these tools to make decisions faster, identify early infections, monitor the condition of the infected patients, provide improved treatment, and develop effective treatments and vaccines. Towards this direction, Covid-19 High-Performance Computing Consortium, a public-private joint effort, has been launched in the US.

In India, things are moving faster now under the National Supercomputing Mission launched in 2015. The mission is going to establish a network of supercomputers ranging from a few teraflops (TF) to hundreds of TFs and systems with greater than or equal to three petaflops (PF) in academic and research institutions by 2022. With three more supercomputers coming up, one each at IIT-Kanpur, JN Centre for Advanced Scientific Research, Bengaluru, and IIT-Hyderabad, the supercomputing facility will be ramped up to six PF. Eleven new systems are planned across India with a cumulative capacity of 10.4PF.
As per medical specialists, Covid-19 is here to stay, and future research in IT will play a vital role in learning, evolving, protecting, and moving towards positive results.
Dr Deepak Halan is associate professor at School of Management Sciences, Apeejay Stya University